Visual Gene Developer
In the previous section, we shortly introduced synthetic gene design. However, gene design in itself may not be easy as it may require computational works to optimize genes and a gene construct may include several different gene elements such as his tag, mRNA hairpin structure, restriction enzyme sites, and conserved Visual Gene Developer in the UTR region.
In order to facilitate gene design, we have been developing unique gene design software called Visual Gene Developer. The software provides a user-friendly interface and includes many useful functions such as codon optimization, mRNA secondary structure prediction, GC content calculation, sequence comparison, repeated sequence search, and restriction enzyme site search. Furthermore, the software supports not only automated gene optimization and network and multi-threaded computing that reduces the processing time significantly but also artificial neural network prediction model. One of the powerful features is that Visual Gene Developer allows users to develop new modules and plug-in with general script languages (VBScript and JScript).
We released the software to the public as freeware. Please check the license agreement.
For more details, check our paper
| Jung, S.-K. and K. McDonald (2011). "Visual Gene Developer: a fully programmable bioinformatics software for synthetic gene optimization." BMC Bioinformatics 12(1): 340 |
o Acknowledgment
* The software development has been partially funded by Chevron Technology Ventures.
* As a former advisor of a software developer, Professor Sun Bok Lee (sblee@postech.ac.kr, http://www.postech.ac.kr/ce/Biotech/) partially contributed to the development of artificial neural network module.
* Dr. Ivo Hofacker (ivo@tbi.univie.ac.at) has shared his mRNA structure prediction program.
* An unknown programmer whose ID is nogChoco in Belgium has shared a free module named 'LineNumbers for the RichTextBox'.
* As a close collaborator, we have worked together with Professor Dandekar (amdandekar@ucdavis.edu)'s group.
o Main window menu
Visual Gene Developer 1.3 (Build 689)

o Screenshots
Startup
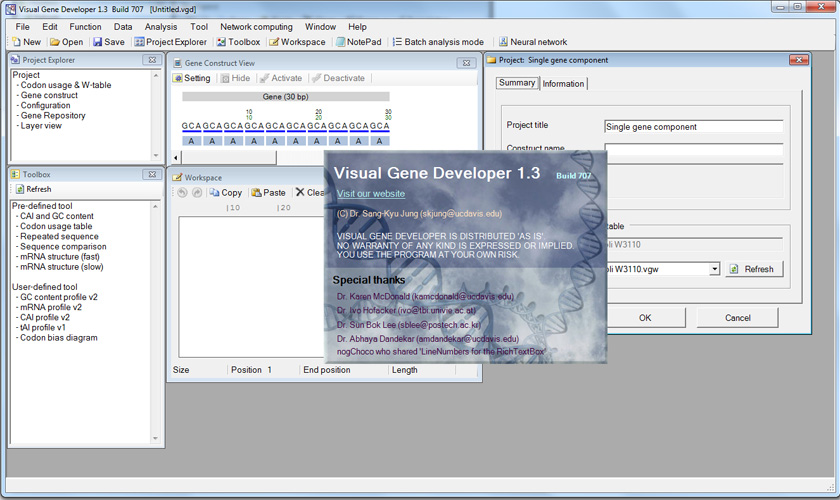
Gene analysis and manipulation
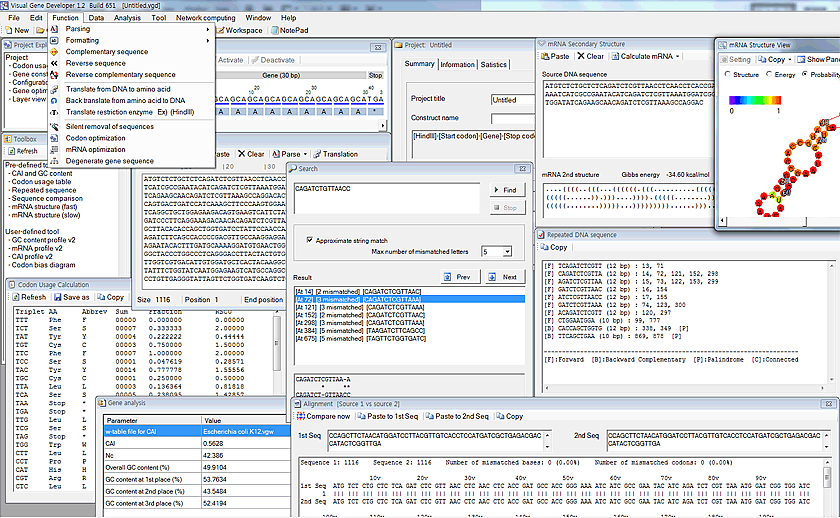
Gene construct design and analysis
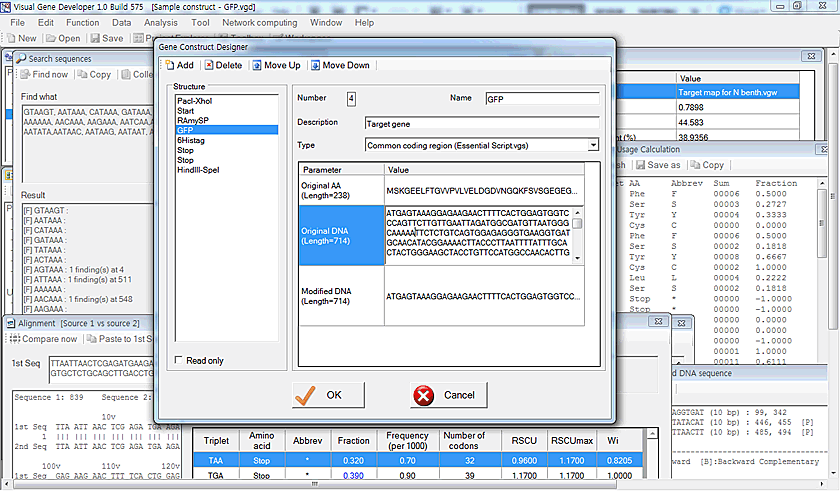
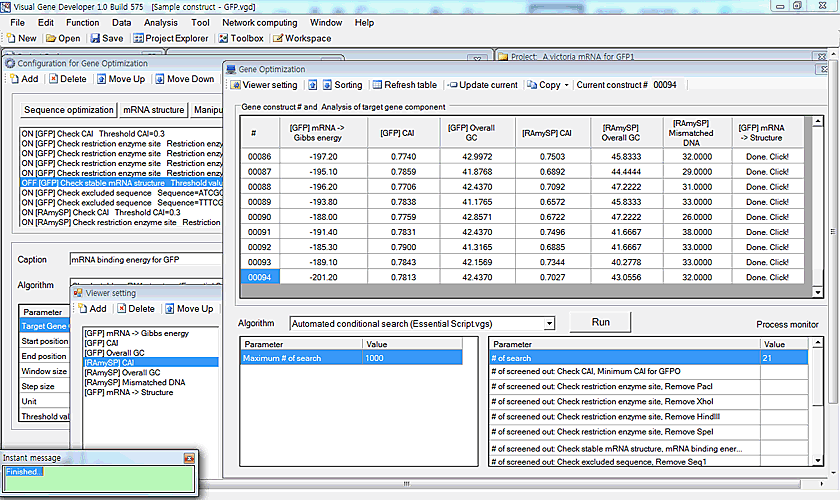
Gene optimization
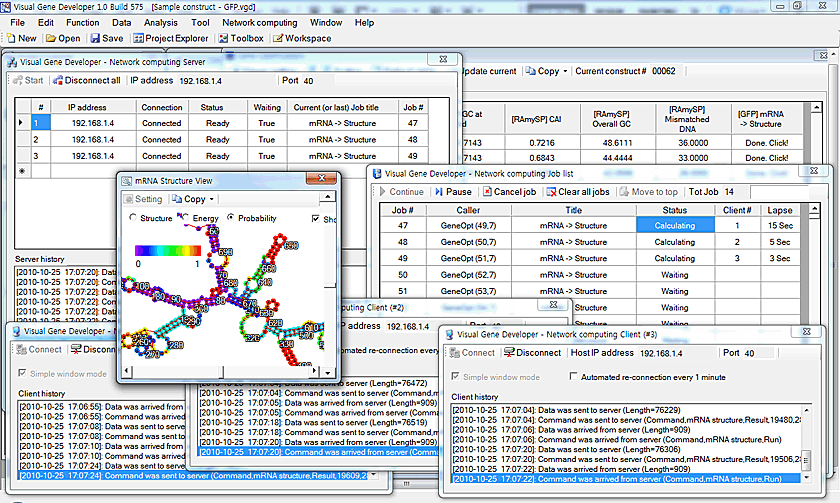
mRNA structure prediction combined with network & multi-threaded computing
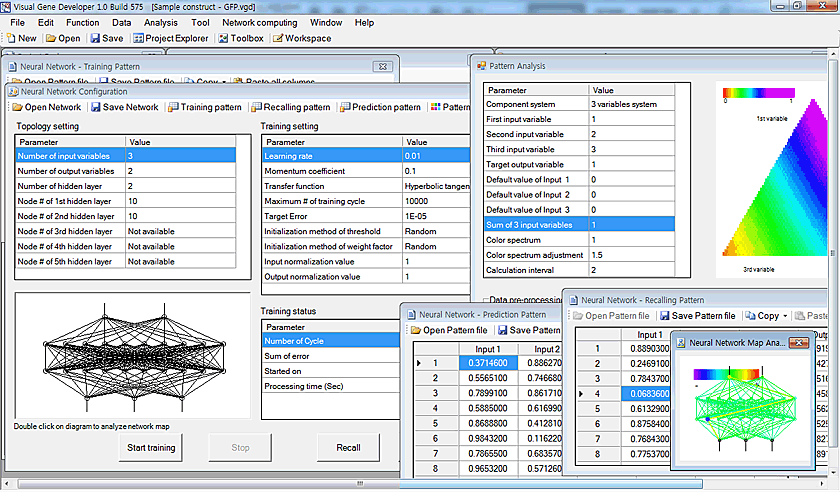
An artificial neural network prediction tool
Module development using a script language